Understanding Translational Effects of Variants With SnpEff
Once we have assembled the genomes of our subject(s)[1][2], generated a list of variants, annotated these variants with relevant databases (e.g. dbSNP)[3], we may now be interested in investigating the structural and translational effects of genomic variants on proteins.
 -Interacting with protein structures in VMD“
-Interacting with protein structures in VMD“
If the interest in structural variations is well-intentioned then it behooves us to use SnpEff, which both adheres to VCF 4.1 standards and GATK best practices. As with many of the downstream processes we must make an initial investment by choosing a reference build, which for human samples, at the moment, consists of GRCh37 and HG19. Install the necessary reference library and run SnpEff:
$java -Xmx[allocate memory] -jar snpEff download [reference library]
$java -Xmx[allocate memory] -jar snpEff eff -v -onlyCoding true -i vcf -o vcf [reference library] [input].vcf > [output]
It should be noted that the choice of reference build is not necessarily arbitrary. The same reference genome that was used for assembly, should have also been used for variant detection, and this rule remains constant for use in uncovering translational effects in SnpEff. Otherwise, the user will be met with a “No Tribble Type” error. Correctly executed, the INFO field of our VCF file will contain the new additional annotations:
SNPEFF_AMINO_ACID_CHANGE=E281*
SNPEFF_CODON_CHANGE=Gag/Tag
SNPEFF_EFFECT=STOP_GAINED
SNPEFF_EXON_ID=NM_032269.ex.6
SNPEFF_FUNCTIONAL_CLASS=NONSENSE
SNPEFF_GENE_BIOTYPE=mRNA
SNPEFF_GENE_NAME=CCDC135
SNPEFF_IMPACT=HIGH
SNPEFF_TRANSCRIPT_ID=NM_152727
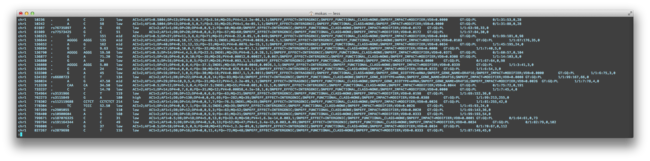
Above we see the fields filled in with a sampling from within the gene CCDC135. Below we can see how this data appears within an intact VCF 4.1 file, which can be parsed to pull out the desired details.
If we choose to create a file containing translation effects which also adheres to GATK best practices, there are a few additional steps, however recent studies have shown that while GATK pipelines are designed to improve results, they don’t always tend to do so[4].
$java -Xmx[allocate memory] -jar GenomeAnalysisTK.jar
-T VariantAnnotator
-R [reference].fasta -A SnpEff --variant [raw].vcf --snpEffFile [snpeffoutput].vcf
-L [raw].vcf -o [gatk_snpeff_output].vcf
The process outlined in this post will bring users closer to understanding how genomic variants cause changes in protein structures and possibly lead to functional insights. Other tools such as SIFT and PolyPhen are also promising in aiding the study of translational changes, investigators are encouraged to compare tools and share opinions. Good luck!